
A few months ago I published a quite popular post on Clustering the Bible… one well known clustering algorithm is k-means. If you want to learn how k-means works and how to apply it in a real-world example, read on…
k-means (not to be confused with k-nearest neighbours or KNN: Teach R to read handwritten Digits with just 4 Lines of Code) is a simple, yet often very effective unsupervised learning algorithm to find similarities in large amounts of data and cluster them accordingly. The hyperparameter k stands for the number of clusters which has to be set beforehand.
The guiding principles are:
- The distance between data points within clusters should be as small as possible.
- The distance of the centroids (= centres of the clusters) should be as big as possible.
Because there are too many possible combinations of all possible clusters comprising all possible data points k-means follows an iterative approach:
- Initialization: assign all data points randomly to k clusters
- E-step (for expectation): assign each point to the nearest (based on Euclidean distance) centroid
- M-step (for maximization): determine new centroids based on the mean of all assigned points
- Repeat steps 2 and 3 until no further changes occur
As can be seen above k-means is an example of a so-called expectation-maximization algorithm.
To implement k-means in R we first assign some variables and define a helper function for plotting the steps:
n <- 3 # no. of centroids
set.seed(1415) # set seed for reproducibility
M1 <- matrix(round(runif(100, 1, 5), 1), ncol = 2)
M2 <- matrix(round(runif(100, 7, 12), 1), ncol = 2)
M3 <- matrix(round(runif(100, 20, 25), 1), ncol = 2)
M <- rbind(M1, M2, M3)
C <- M[1:n, ] # define centroids as first n objects
obs <- length(M) / 2
A <- sample(1:n, obs, replace = TRUE) # assign objects to centroids at random
colors <- seq(10, 200, 25)
clusterplot <- function(M, C, txt) {
plot(M, main = txt, xlab = "", ylab = "")
for(i in 1:n) {
points(C[i, , drop = FALSE], pch = 23, lwd = 3, col = colors[i])
points(M[A == i, , drop = FALSE], col = colors[i])
}
}
clusterplot(M, C, "Initialization")
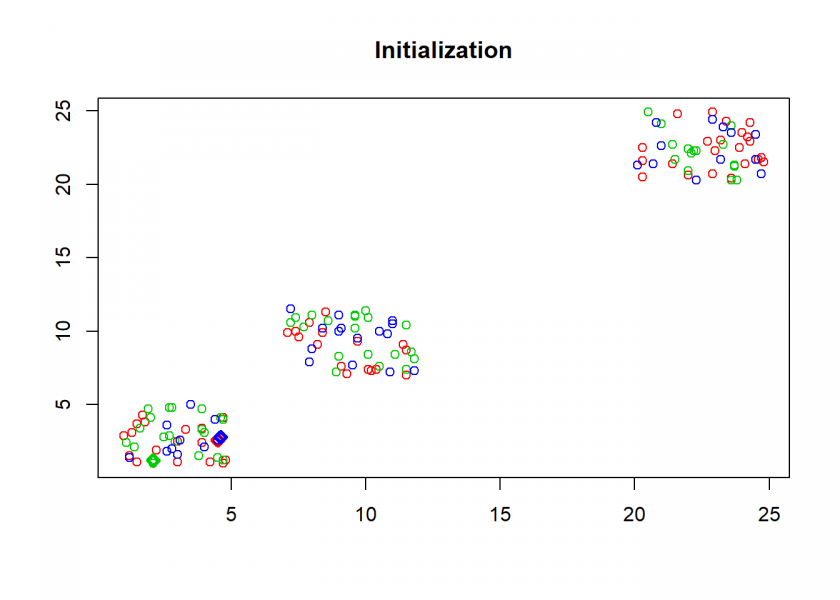
Here comes the k-means algorithm as described above (the circles are the data points, diamonds are the centroids and the three colours symbolize cluster assignments):
repeat {
# calculate Euclidean distance between objects and centroids
D <- matrix(data = NA, nrow = n, ncol = obs)
for(i in 1:n) {
for(j in 1:obs) {
D[i, j] <- sqrt((M[j, 1] - C[i, 1])^2 + (M[j, 2] - C[i, 2])^2)
}
}
O <- A
## E-step: parameters are fixed, distributions are optimized
A <- max.col(t(-D)) # assign objects to centroids
if(all(O == A)) break # if no change stop
clusterplot(M, C, "E-step")
## M-step: distributions are fixed, parameters are optimized
# determine new centroids based on mean of assigned objects
for(i in 1:n) {
C[i, ] <- apply(M[A == i, , drop = FALSE], 2, mean)
}
clusterplot(M, C, "M-step")
}
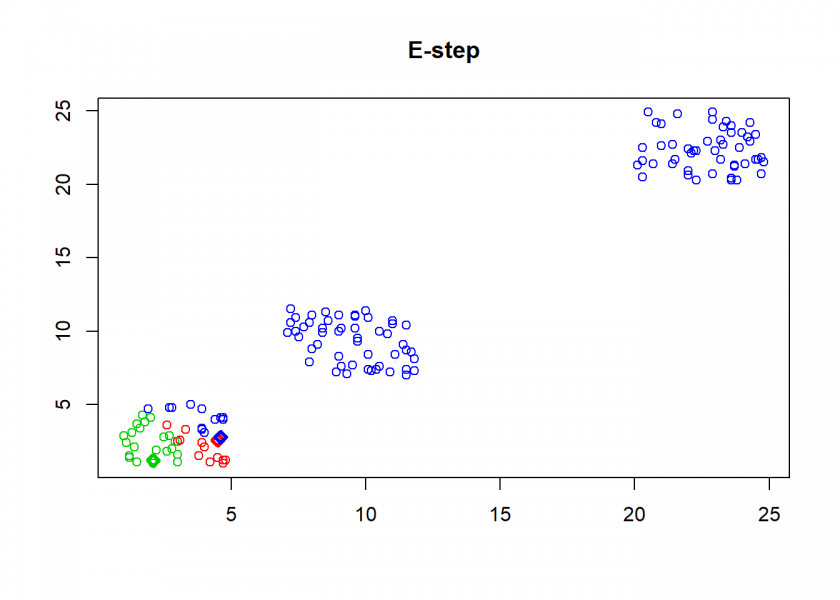
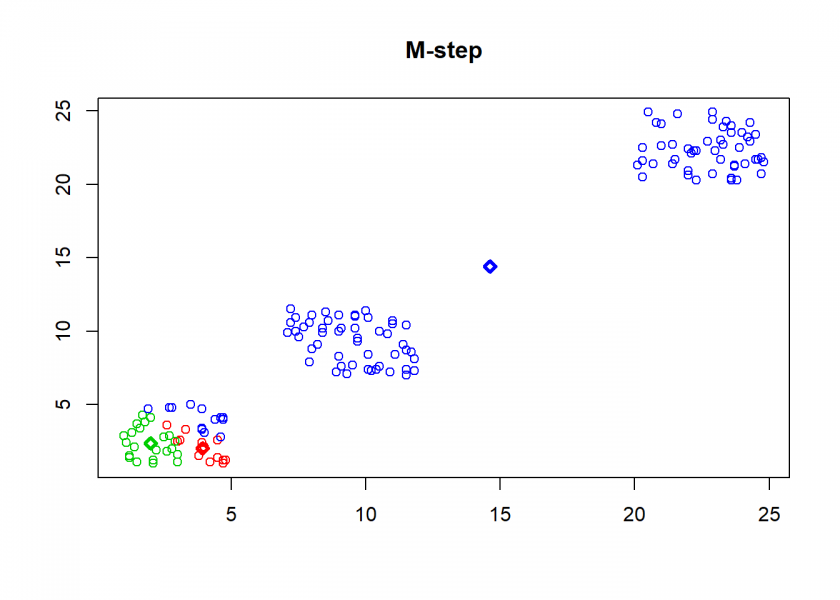
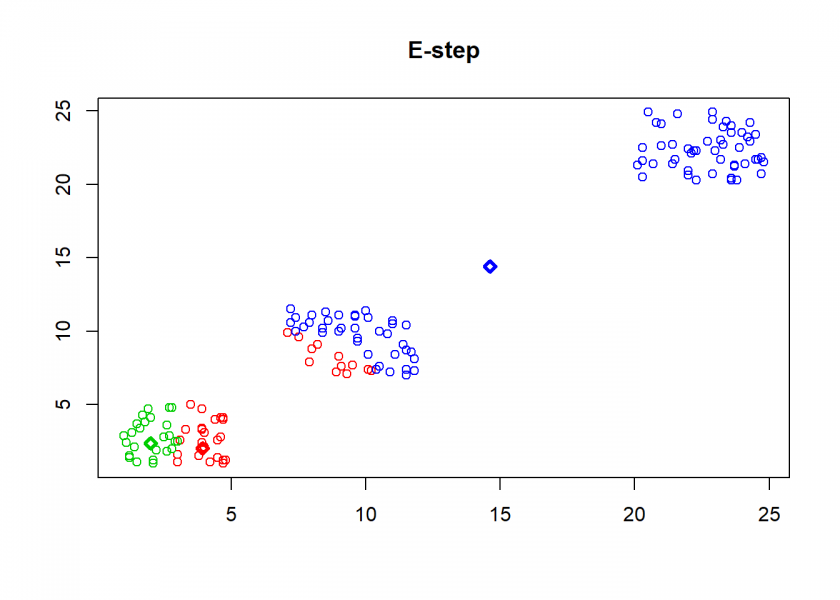
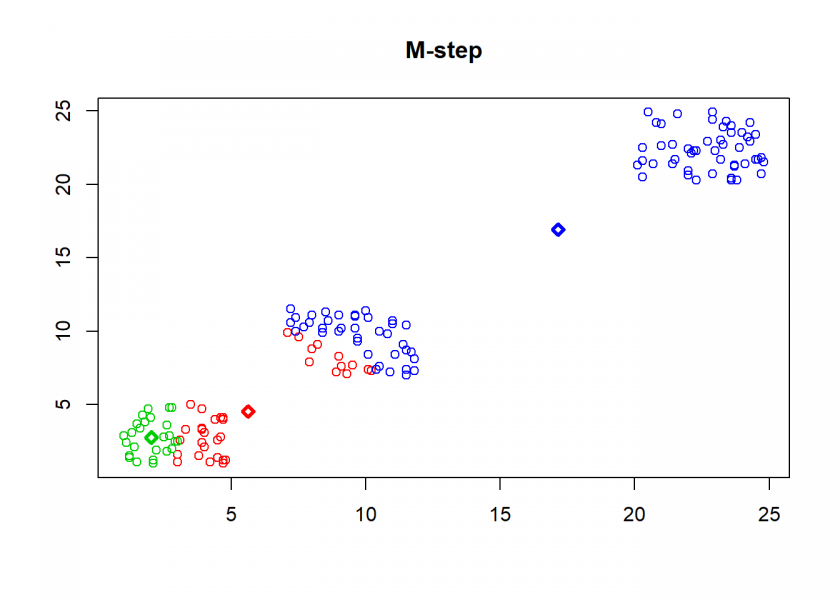
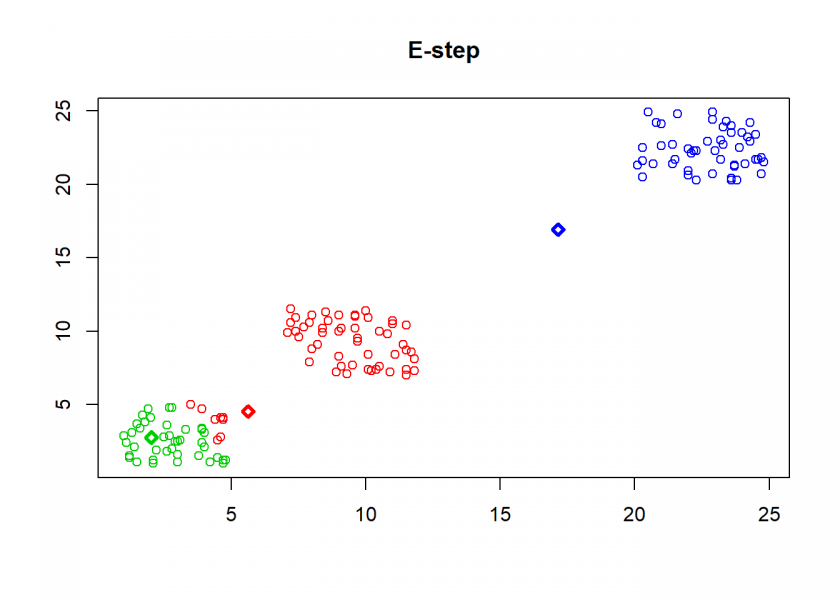
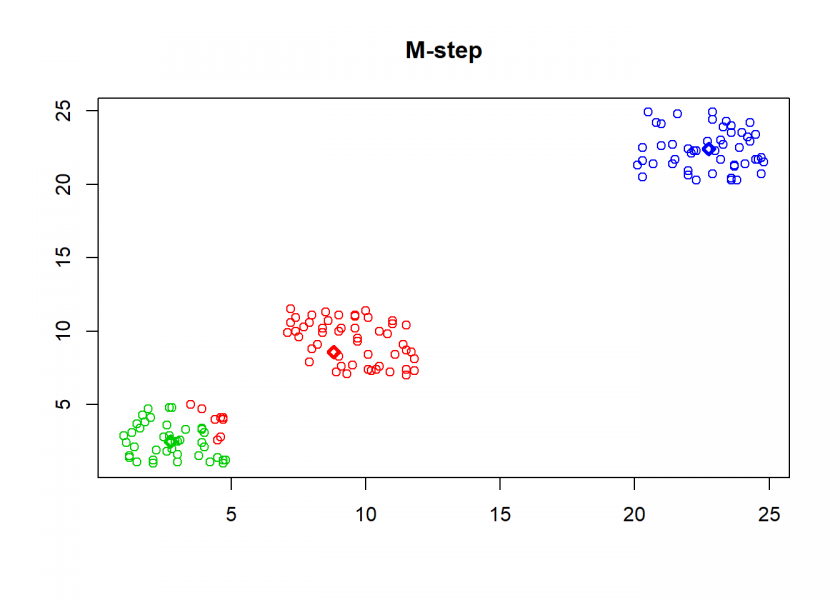
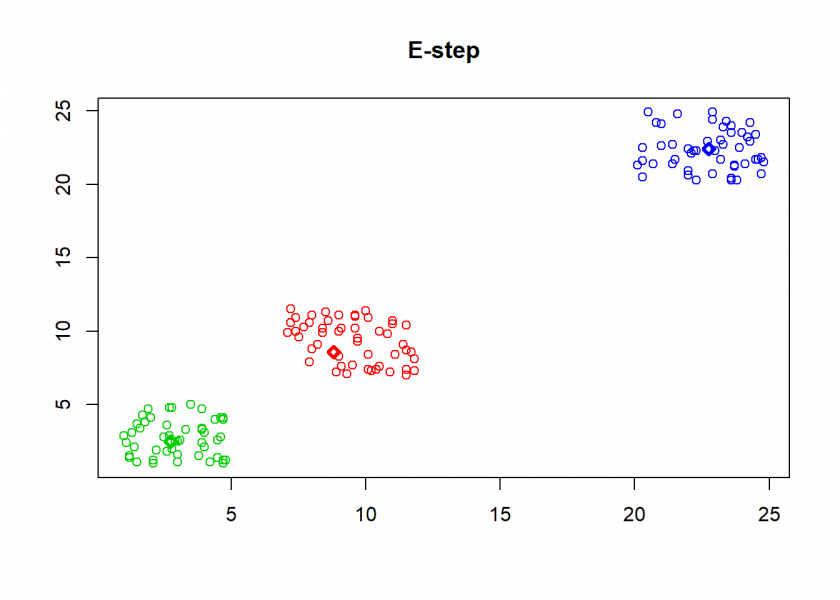
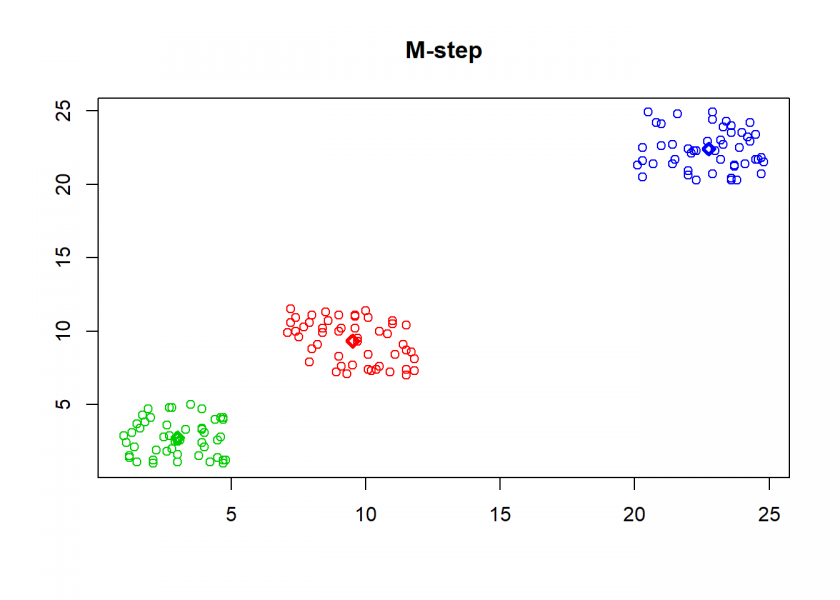
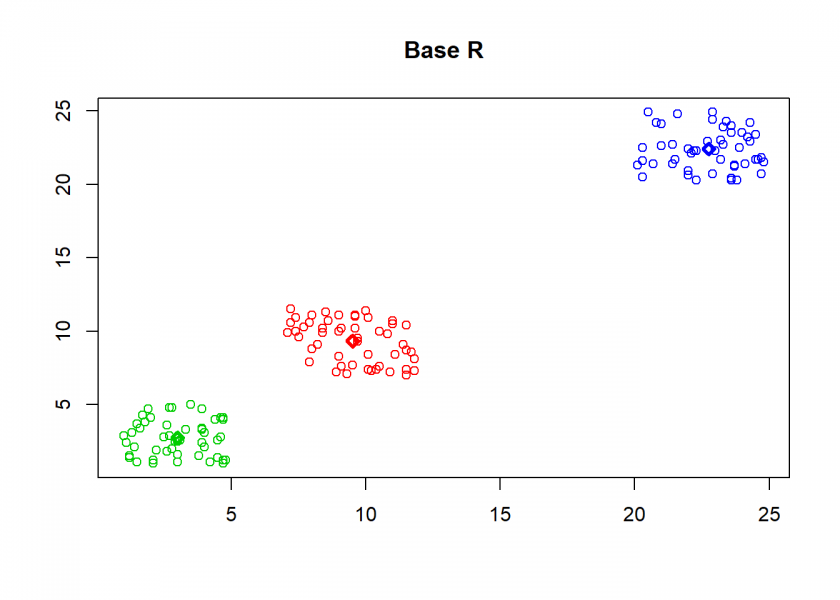
As can seen the clusters wander slowly but surely until all three are stable. We now compare the result with the kmeans function in Base R:
cl <- kmeans(M, n) clusterplot(M, cl$centers, "Base R")
(custom <- C[order(C[ , 1]), ]) ## [,1] [,2] ## [1,] 3.008 2.740 ## [2,] 9.518 9.326 ## [3,] 22.754 22.396 (base <- cl$centers[order(cl$centers[ , 1]), ]) ## [,1] [,2] ## 2 3.008 2.740 ## 1 9.518 9.326 ## 3 22.754 22.396 round(base - custom, 13) ## [,1] [,2] ## 2 0 0 ## 1 0 0 ## 3 0 0
As you can see, the result is the same!
Now, for some real-world application: clustering wholesale customer data. The data set refers to clients of a wholesale distributor. It includes the annual spending on diverse product categories and is from the renowned UCI Machine Learning Repository (I guess the category “Delicassen” should rather be “Delicatessen”).
Have a look at the following code:
data <- read.csv("https://archive.ics.uci.edu/ml/machine-learning-databases/00292/Wholesale customers data.csv", header = TRUE)
head(data)
## Channel Region Fresh Milk Grocery Frozen Detergents_Paper Delicassen
## 1 2 3 12669 9656 7561 214 2674 1338
## 2 2 3 7057 9810 9568 1762 3293 1776
## 3 2 3 6353 8808 7684 2405 3516 7844
## 4 1 3 13265 1196 4221 6404 507 1788
## 5 2 3 22615 5410 7198 3915 1777 5185
## 6 2 3 9413 8259 5126 666 1795 1451
set.seed(123)
k <- kmeans(data[ , -c(1, 2)], centers = 4) # remove columns 1 and 2, create 4 clusters
(centers <- k$centers) # display cluster centers
## Fresh Milk Grocery Frozen Detergents_Paper Delicassen
## 1 8149.837 18715.857 27756.592 2034.714 12523.020 2282.143
## 2 20598.389 3789.425 5027.274 3993.540 1120.142 1638.398
## 3 48777.375 6607.375 6197.792 9462.792 932.125 4435.333
## 4 5442.969 4120.071 5597.087 2258.157 1989.299 1053.272
round(prop.table(centers, 2) * 100) # percentage of sales per category
## Fresh Milk Grocery Frozen Detergents_Paper Delicassen
## 1 10 56 62 11 76 24
## 2 25 11 11 22 7 17
## 3 59 20 14 53 6 47
## 4 7 12 13 13 12 11
table(k$cluster) # number of customers per cluster
##
## 1 2 3 4
## 49 113 24 254
One interpretation could be the following for the four clusters:
- Big general shops
- Small food shops
- Big food shops
- Small general shops
As you can see, the interpretation of some clusters found by the algorithm can be quite a challenge. If you have a better idea of how to interpret the result please tell me in the comments below!
Setting ‘k’ = supervised…
Cheers,
Andrej
Dear Andrej,
Supervised means that one attribute of your data is the target variable, with clustering all data are equal, therefore it is un-supervised!
k is a so called hyperparamter which specifies the number of clusters.
Hope that helps!
It’s possible to estimate (or “learn” as machine learning people would say) K as well as the clusters using a Dirichlet process (or other “non-parametric” prior). With K-means, the y[n] in R^d are the d-dimensional vectors you’re clustering. The missing data is the cluster z[n] in 1:K to which they’re assigned. If you knew that, this would reduce to training a normal classifier.
There’s no penalty in the K-means model for centroids being closed together and there’s no attempt in the algorithm to keep them far apart. If there was, it’d show up as a penalty term or prior in the model. An algorithm called K-means++ tries to initialize centroids far from each other with such a penalty to avoid climbing to a bad local maximum.
Theoretically, simple K-means as presented here can be viewed as a Gaussian (normal) mixture model with unit covariance. As such, you can use the expectation maximization (EM) to fit it. The expectation (E) step computes the expectations, which here reduce to the probability a point is in each cluster, then use those weights to do a maximization (M) step (using what an ML person would call “weighted” training).
The algorithm that’s most often called “K-means” is the one you present. It’s a greedy heuristic strategy for hill climbing to a local maximum. Finding the true maximum likelihood cluster assignment is NP-hard due to the combinatorics and almost never attempted as N and K are usually prohibitively large for exponential search. So the answer depends on the initialization. If you rerun K-means with new initial centers, you’ll get a different result on most non-trivial data sets. The EM algorithm will also get stuck in a local maximum, just like the simpler greedy algorithm.
Thank you very much for this instructive and elaborate comment, Bob!
I usually run a ranked cross-correlation into my dataset to start understanding the clusters:
library(lares) #devtools::install_github(“laresbernardo/lares”)
clusters <- clusterKmeans(df, k = 5) # df is any data.frame. One hot smart enconding will be applied if categorical values are present
clusters$correlations # Same as corr_cross(clusters$df, contains = "cluster")
Hope it comes helpful 😉
Thank you, very interesting. Are you planning to publish the package on CRAN?
I see it more like an everyday package than like a library. It has too many useful and various kinds of functions that I have gladly shared with the community, but I think CRAN is not aiming for this kind of libraries in their repertoire! But thanks appreciate your comment though. Feel free to dive into the lares package for more useful functions 😉